Intro
For a long time, Pandas was, and still is arguably the single most important library inside a data scientist's toolbox. In fact, it was the emergence of Pandas that helped to make Python such a popular programming language, both for data science tasks and generally. But datasets continue to grow, and now they often exceed the size of available RAM on most machines. Thus, the need for fast and efficient processing and analysis of data, i.e. the next generation of tooling grows too.
There are several popular technologies in the Python ecosystem that are frequently used for processing large datasets in the context of data science and data engineering. Dask is an open-source, general framework for parallel and distributed computations in Python. It is often the go-to technology for horizontal scaling of various types of computations and data science tasks. Its higher level API, the Dask.DataFrame provides arguably the most convenient way to sidestep the memory and performance hurdles one can encounter with Pandas when working even with moderately large datasets.
Vaex is a high-performance DataFrame library in Python, primarily built for the processing, exploration and analysis of datasets as large as the size of your hard-drive and on a single machine. When it comes to large datasets, Vaex argues that horizontal scaling, i.e. adding more computers/nodes, is simply not needed for the majority of the common data science and analytics tasks, provided one uses the right approaches for storing, accessing, and processing of the data.
To a user that is generally aware but not intimately familiar with one or both of these technologies, they may seem quite similar to each other. Both are often used to replace Pandas when performance or memory management becomes an issue. Both use concepts like lazy (or delayed) evaluations, parallelize computations and employ out-of-core algorithms. On top of this, Dask can distribute computations on a cluster. So one might wonder: "What is the real difference between these two libraries?" or "When or why would I want to use Vaex"? In what follows I will highlight the main differences between these two technologies, hoping to improve your understanding of them, and enable you to make a more informed choice when choosing the right tools for your use-case.
1. Dask vs. Dask.DataFrame vs. Vaex
Dask is a general purpose framework for parallelizing or distributing various computations on a cluster. A number of popular data science libraries such as scikit-learn, XGBoost, xarray, Perfect and others may use Dask to parallelize or distribute their computations.
When many data scientists or engineers say "Dask" what they often mean is Dask.DataFrame. Dask.DataFrame is a DataFrame library built on top of Pandas and Dask. Essentially a Dask.DataFrame is composed of many smaller Pandas DataFrames which are coupled to a generic task scheduler provided by Dask. When a computation is triggered, Dask will break up the Dask.DataFrame into many smaller Pandas DataFrames, each doing their part of the work, and join the result at end.
Vaex is a fully new DataFrame implementation, built from the ground up to work incredibly fast with DataFrames comprising hundreds of millions or billions of rows. While it tries to follow the API set by Pandas, under the hood lie efficient, out-of-core C++ algorithms. Coupled with memory-mapping, it can process ~1.000.000.000 rows in a second or two. Vaex can also stream data to and from your favorite cloud storage buckets, and is fully compatible with Apache Arrow, allowing you to receive and share data with eco-systems outside of the Python realm.
2. Lazy evaluation
Both Dask.DataFrame and Vaex support lazy (or delayed) evaluations. There are some differences in how they go about it however, that the user should be aware of. In Dask.DataFrame, any operation that requires iterating over the data is delayed until the user explicitly specifies that the calculation should be done by invoking the .compute() method. For example, calculating the mean of a column would look like this:
from dask import dataframe as dd
dask_df = dd.read_parquet('example.parquet')
mean_x = dask_df.x.mean().compute()
print(mean_x)If .compute() is not specified in the above example, mean_x will be just a placeholder waiting to house the result once the computation is triggered. Even for "simple" operations such as getting the shape of a DataFrame, one is expected to invoke the .compute() method :
from dask import dataframe as dd
dask_df = dd.read_parquet('example.parquet')
shape = dask_df.shape
print(shape[0].compute(), shape[1])This makes sense: the first dimension of the .shape method specifies the number of rows, which in theory can be quite large and requires Dask.DataFrame to iterate over the entire data to count it. The second dimension is the number of columns, which can be considered as "metadata" and is obtained immediately. The advantage of this approach is that allows for some advanced optimization techniques, since the entire computational graph is known prior to execution. For example:
mean_x = dask_df.x.mean()
sum_x = dask_df.x.sum()
result = dd.compute(mean_x, sum_x)
print(result)In the above code block the mean and the sum of x are both calculated with one pass over the data.
Vaex handles the lazy evaluations differently. If a result of an operation will "live" inside of the DataFrame from which it was defined, the operation will be delayed. An example of this is adding a new column, or joining two DataFrames together. On the other hand, if a result of an operation will live "outside" of the DataFrame from which it was defined, the operation will be triggered immediately. An example of this is calculating the mean of a column, finding the unique values of a column, or a groupby operation. For example:
import vaex
vaex_df = vaex.open('example.parquet')
# Column r is added lazily
vaex_df['r'] = (vaex_df['x']**2 + vaex_df['y']**2)**0.5
# Mean of r will be calculated eagerly
mean_r = vaex_df.r.mean()
print(mean_r)A workflow such as this is quite convenient to many data scientists which require immediate feedback especially in the often interactive data exploration and analysis stages. While computing multiple aggregations or calculations with one pass over the data is also possible with Vaex, the API of Dask is more convenient for this approach.
3. Handling large datasets
Being able to handle large amounts of data is a common reason for using either of these two libraries. Their approach to handling such data is a bit different however. Dask.DataFrame overcomes this challenge by chunking the data into multiple Pandas DataFrames which are then lazily evaluated. Dask.DataFrame can also distribute these smaller, more manageable DataFrames on a cluster. The upside here is that one can use a cluster of arbitrary size to overcome any memory issues, and parallelize the computations. Dask.DataFrame follows more closely the Pandas API compared to Vaex, and provides a greater coverage of the Pandas functionality. The downside stems from the same reason - Dask.DataFrame inherits some of the baggage of Pandas, such as being memory hungry. Also introducing a cluster into a data science workflow introduces a significant complexity related to its setup, management, development, and costs. Organizations such as Coiled and SaturnCloud are trying to simplify this challenge.
Vaex completely rethinks how a modern DataFrame should be implemented. It all starts with the data, which should be in a memory-mappable format on disk. Coupled with the read speed of ~5GB/s for modern SSD disks and ~12.5GB/s for cloud buckets allows the data to be read and processed with incredible speed - all on a single machine, even a laptop! While a performance comparison is beyond the scope of this article, check out how fast Vaex can do some common operations on over 1.000.000.000 (billion) samples:
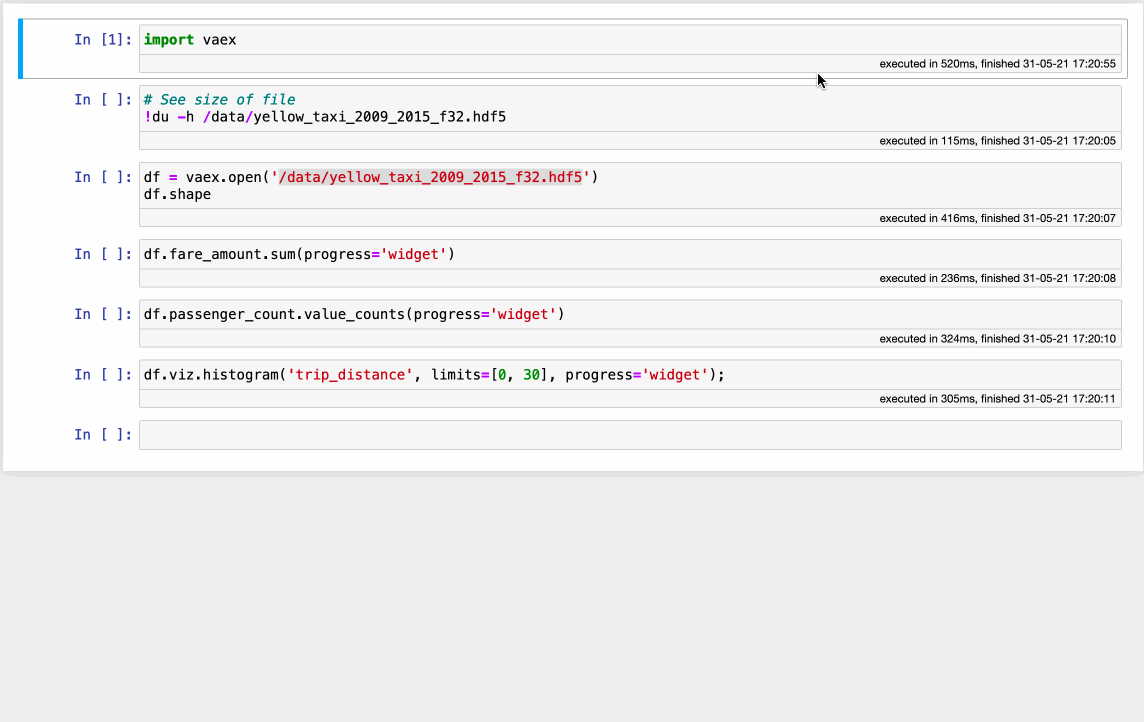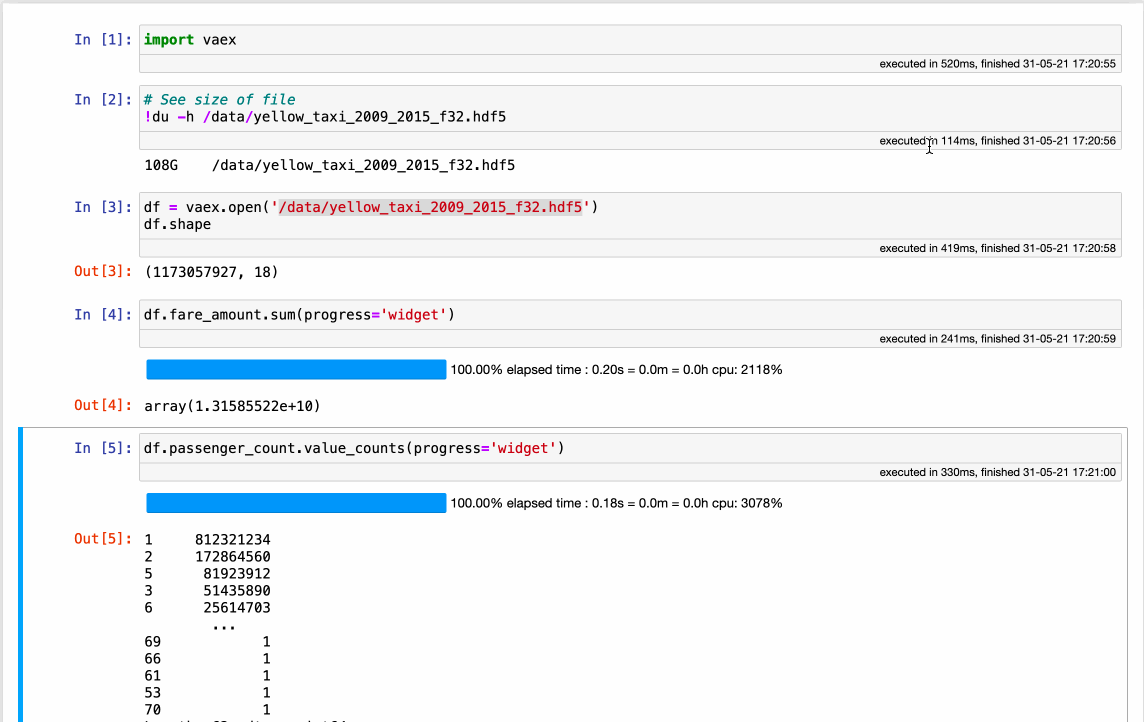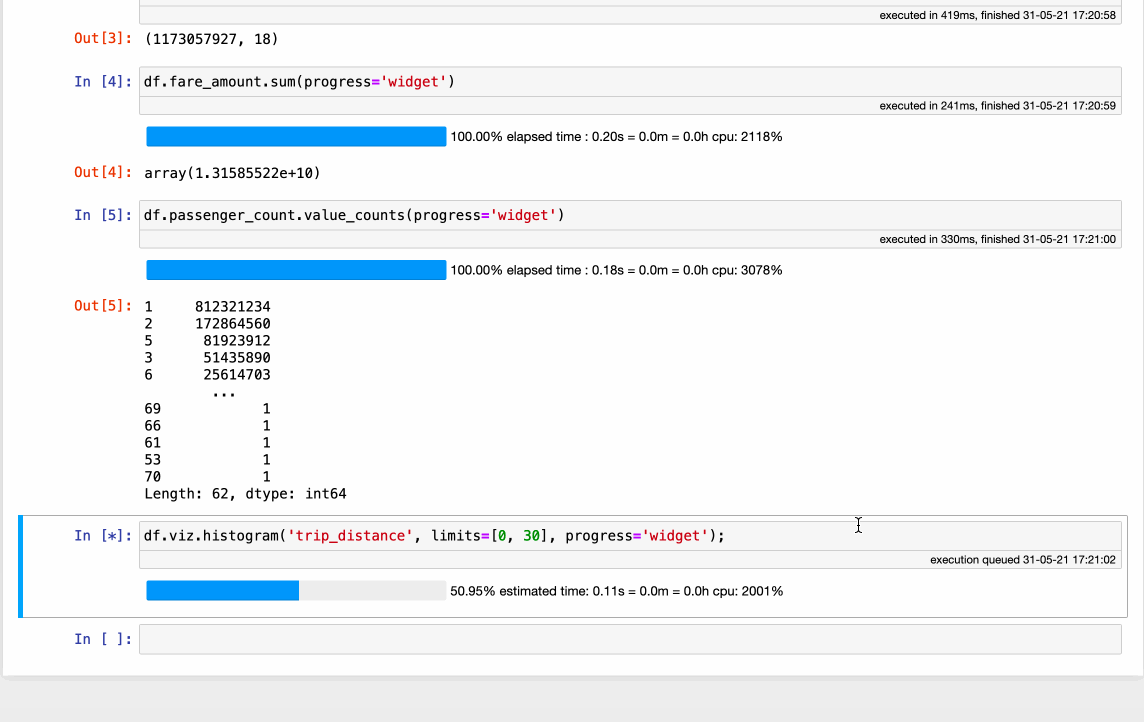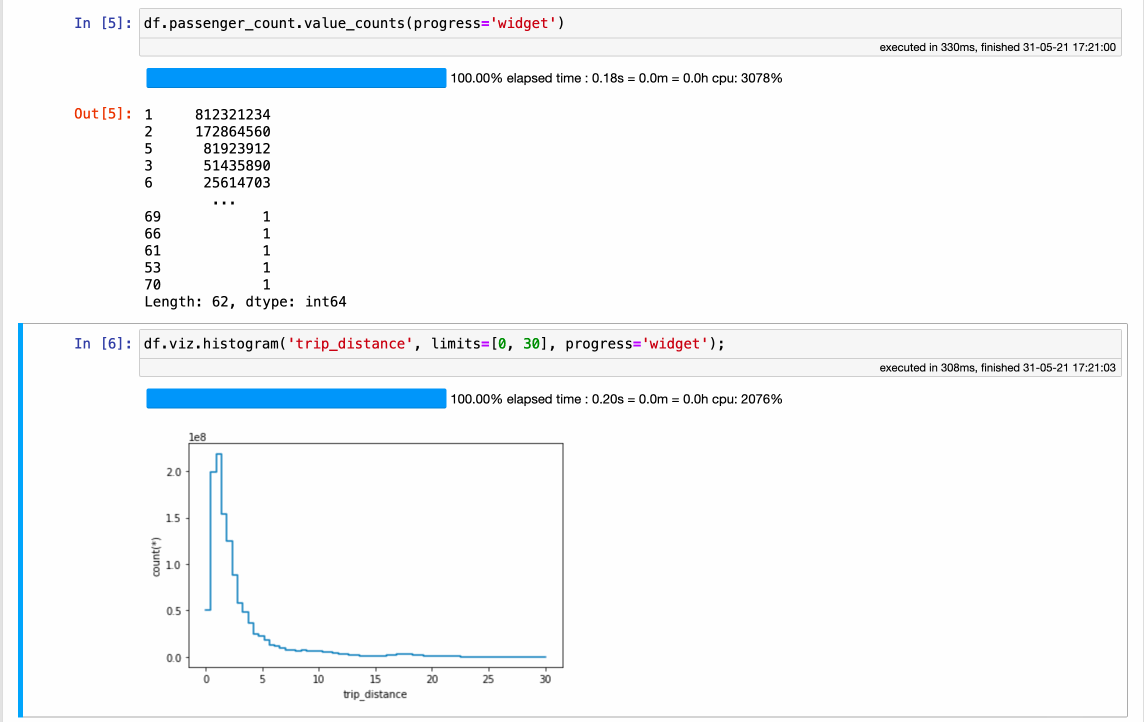
The obvious benefit of using Vaex is the incredible out-of-the-box speed, with absolutely no setup or need for any special system configuration or cluster provisioning. The downside is that, since this is a new implementation, it does not yet have all methods that Pandas offers.
While any kind of performance comparison is outside the scope of this article, if you are interested you should check out this post. All benchmarks are biased in one way or another, so I would encourage you to try everything yourself, using your data, and judge relative to your needs.
4. Data file formats
Apache Parquet is the preferred data format of Dask.DataFrame. For larger datasets, one gets better performance if the data is partitioned amongst multiple files. When using Dask.DataFrame on a cluster, the data can be centralized i.e. located on a single machine, or distributed among the machines comprising the cluster.
For best performance, the preferred data formats of Vaex are HDF5 and Apache Arrow. Vaex can also read the Apache Parquet file format. Vaex expects all of the data to be located on a single machine. While it can lazily read datasets that are spread amongst multiple files, for optimal performance it is best that the entire data is stored in a single file. Like Dask, Vaex can also stream data from (and to!) your favorite Cloud object storage, and optionally cache it locally.
Both libraries can read data stored in CSV, JSON, and virtually any file format that Pandas can read. However, for larger datasets and better performance, it is better to convert the data to a file format preferred by each of the libraries.
5. What to use
It is difficult to say anything in this section here that is completely unbiased. I would kindly but strongly recommend to you, dear reader, to try these and any other library yourself, and make decisions relative to your data, use-cases and needs. With that in mind, the following is my personal, biased take.
Vaex is flat out the fastest and most memory efficient Python DataFrame library out there. The fact that it can process 1.000.000.000 (billion) rows per second on a single machine is unmatched in the industry today. It is made for interactive exploration, visualization, and preprocessing of large tabular datasets. Being a single machine solution, it adds virtually zero DevOps overhead, and the stunning performance makes it an excellent backend engine for a variety of interactive dashboards or machine learning applications.
There are still plenty of computational tasks in data science and even more in data engineering which can not be done in an out-of-core manner. Parsing a very large number of xml files, for an example. This is where Dask shines. Since it is a general framework for distributed computing, it can generalize well outside the domain of tabular datasets. Dask.DataFrame the DataFrame implementation built on top of Dask and Pandas, provides a much more complete API spec relative to Pandas. It also provides a way to parallelize and distribute a variety of common machine learning operations that are often done with popular Python libraries such as Scikit-Learn or XGBoost.
Outro
Dask is a general purpose tool, and takes a general purpose approach to scaling Pandas. It builds a large DataFrame out of small Pandas DataFrames, coupled to a generic task scheduling framework. This provides broad reach while leaving room for optimization. On the other hand, Vaex is more specialized around the large DataFrame problem. This allows it to be more efficient, at the cost of some generalizability.
Vaex and Dask[.DataFrame] are both excellent libraries for data science. While this article focused on comparing and contrasting their approaches to processing large tabular datasets, I can think of a variety of use-cases where an optimal solution would employ both technologies.
These two libraries are just a small testament to the strength of the Python ecosystem, and together with the many other libraries out there ensure that we can tackle any problem that we encounter. The fact that there are multiple ways to attack a particular problem can only be a good thing!
Thanks to Matthew Rocklin for providing helpful comments and suggestions that improved this article.
